2016 DIMACS REU Proposed Project Descriptions
*Note that you must be a U.S. Citizen or Permanent Resident to be eligible for these projects.
Project #: DC2016-01
Mentor: Eric Allender, PhD, Computer Science, Rutgers UniversityProject Title: The Minimum Circuit Size Problem: A possible NP-Intermediate Problem
The Minimum Circuit Size Problem (MCSP) is a computational problem in NP that is widely believed to be hard to compute, although many researchers suspect that it is NOT NP-complete. However, there is currently only weak evidence to support this suspicion. This REU project will endeavor to find stronger evidence, of the form "If MCSP is NP-complete, then something weird happens". Another possible direction involves clarifying the relative complexity of MCSP and other candidate NP-Intermediate problems. (The PI has previously published research with an REU student on this topic, and there is more work to be done, which should be accessible to an REU student.)
References:
Eric Allender, Dhiraj Holden, Valentine Kabanets: The Minimum Oracle Circuit Size Problem. STACS 2015: 21-33
Eric Allender, Bireswar Das: Zero Knowledge and Circuit Minimization. MFCS (2) 2014: 25-32
Cody D. Murray, Richard Ryan Williams: On the (Non) NP-Hardness of Computing Circuit Complexity. Conference on Computational Complexity 2015: 365-380
Project #: DC2016-02
Mentor: Periklis Papakonstantinou, Management Science and Information Systems, Rutgers UniversityProject Title: Multi-stream algorithms: design and analysis
The streaming model is the prototypical mathematical formalization for analyzing Massive Datasets. In the classical streaming model the data elements arrive one-by-one for a processor to compute. This processor is limited to use a tiny small local memory (compared to the total number of elements), where it keeps track of what happened. Typically, the memory size is poly-logarithmic in the stream size. Surprisingly, there are many interesting problems that can be solved in the streaming model.
In 2009 (see below [1]) a seminal research article introduced the multi-stream model and initiated the investigation of its limitations. The multi-stream model uses two (or more streams), which the algorithm can scan a very small number of times, e.g. 2 or 3. Each stream can be thought to correspond to a hard disk-array. Thus, the study of the multi-stream model blends perfectly theory and practice.
There are several important works studying limitations of multi-stream algorithms. In contrast, this project proposes to study algorithms (practical constructions). Some first algorithmic techniques were developed in an article [2] published a year ago, which regards cryptographic constructions in the multi-stream model. Now, we propose the study of more elementary combinatorial problems and algorithms. These problems include computation and approximation of linear algebra problems.
This project does not require (neither will involve) knowledge in cryptography. It does require though some level of mathematical maturity and good knowledge of basic probability, linear algebra, and combinatorics, and excitement for algorithms and their modern applications as in e.g. computation over big data.
References:1. Martin Grohe, André Hernich, Nicole Schweikardt: Lower bounds for processing data with few random accesses to external memory. Journal of ACM, 2009
2. Periklis A. Papakonstantinou, Guang Yang Cryptography with Streaming Algorithms. CRYPTO, 2014
Project #: DC2016-03
Mentor: Periklis Papakonstantinou, Management Science and Information Systems, Rutgers UniversityProject Title: Learning streaming algorithms
We are given a function f described on a few points through a number of examples {(f(x),x) | x: an input}. Furthermore, we have a promise that the problem f can be computed by an algorithm able to deal with massive streams of data, i.e. when each x is really big. The question is to do Machine Learning on Big Data, that is to construct the program of the streaming algorithm from the given examples (such that the same algorithm will work correct or approximately correct on unseen inputs x). A streaming algorithm is one that scans its input from left-to-right once or a small number of times and at the same time is restricted to use a very small local memory (say of size logn or poly-logn, where |x|=n). This general task is known to be impossible in its generality (in fact, even for automata/i.e. steaming algorithms with constant local memory). This project sets a very exciting goal of re-asking the question in the "correct" context and checking whether under certain natural restrictions learning streaming algorithms can be done.
This project assumes no prior knowledge in machine learning, but elements of ML will be studied during the project. It does require some level of mathematical maturity and good knowledge of basic probability and combinatorics. Excitement for algorithms and their modern applications, as in e.g. computation over big data and machine learning, is necessary.
Project #: DC2016-04
Mentor: Chang Chan, Rutgers Cancer Institute of New JerseyProject Title: Data Mining of Genomic Information from Large Cancer Data Sets
Very large data sets of genomic information on cancer including somatic mutations and gene expressions of tumors from diverse cancer types have been collected and continues to grow. There are many opportunities to mine these data sets for patterns across tumor types as well as those specific to a tumor type. The methods will include data visualization, statistical methods, and machine learning.
Project #: DC2016-05
Mentor: Weihong ‘Grace’ Guo, Industrial and Systems Engineering, Rutgers UniversityProject Title: Complexity Modeling in Multi-stage, Mixed Model Assembly Systems
Product variety induced manufacturing complexity is a critical problem faced by all manufacturers in today’s competitive environment as manufacturers are developing customized products to respond to individual needs. This study will develop mathematical models on product variety induced manufacturing complexity in multi-stage, mixed model assembly systems. Bayesian network (BN) models and algebraic operators will be developed to obtain key system performance metrics such as process cycle times, operational states of a system, quality, reliability, etc., and to quantify the impact of the complexity on the system performance. The algebraic expressions will be mathematically formalized into algebraic fields and thus provide a way to solve problems such as finding bottleneck stations and achieving line balancing for a complicated production system.
Requirements: basic statistics, math, linear algebra, Matlab
References:
Andres G. Abad, Weihong Guo & Jionghua Jin (2014) Algebraic expression of system configurations and performance metrics for mixed-model assembly systems, IIE Transactions, 46:3, 230-248, DOI: 10.1080/0740817X.2013.813093
Project #: DC2016-06
Mentor: Weihong ‘Grace’ Guo, Industrial and Systems Engineering, Rutgers UniversityProject Title: Sensor Fusion for Process Monitoring and Fault Diagnosis
The wide applications of low-cost and smart sensing devices along with fast and advanced computer systems have resulted in a rich data environment, which makes a large amount of data available in many applications. Sensor signals acquired during the process contain rich information that can be used to facilitate effective monitoring of operational quality, early detection of system anomalies, quick diagnosis of fault root causes, and intelligent system design and control. In discrete manufacturing and many other applications, the sensor measurements provided by online sensing and data capturing technology are time- or spatial-dependent functional data, also called profile data. In this study we are developing methods for effective monitoring and diagnosis of multi-sensor heterogeneous profile data. The proposed method extracts features from the original profile data and then feeds the features into classifiers to detect faulty operations and recognize fault types. Extensive Matlab simulations will be needed to validate the method and explore properties of the proposed method.
Requirements: statistics, math, linear algebra, Matlab
References:
Woodall, W.H. (2007) Current Research on Profile Monitoring. Produção, 17, 420-425.
Noorossana, R., Saghaei, A and Amiri, A. (2012) Statistical Analysis of Profile Monitoring. New York: Wiley.
Lu, H., Plataniotis, K.N. and Venetsanopoulos, A.N. (2009) Uncorrelated Multilinear Discriminant Analysis With Regularization and Aggregation For Tensor Object Recognition. IEEE Transactions on Neural Networks, 20, 103-123.
Project #: DC2016-07
Mentor: Gyan Bhanot, Rutgers Cancer Institute of New JerseyProject Title: Identifying Networks associated with Barretts Esophagus progression to Esophageal Cancer in the BAR-T cell model
Barretts esophagus is a pre-malignant condition found in people with GIRD (Gastro Intestinal Reflux Disease). Using RNA-seq data collected from immortalized Barretts cells treated with acid and bile for 70 weeks, the project involves identifying networks of genes which are disregulated as a function of time. The goal is to identify and study the gene networks in this model which are responsible for malignant transformation of Barretts cells into tumorigenic cells.
Project #: DC2016-08
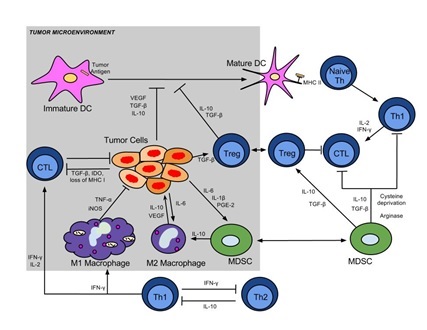
Mentor: Ned Lattime & Chang Chan, Rutgers Cancer Institute of New JerseyProject Title: Whole Transcriptome Sequencing and Analyzing the Immune Tumor Microenvironment
The overall goal of studies in our laboratory spanning preclinical animal models and clinical trials is the analysis of the tumor-immune interaction. Our studies involve the study of oncolytic viral vectors and vaccines and their effects in regulating host immunity with the ultimate goal of inducing an effective tumor specific immune response that will have activity against the primary tumor and metastastatic disease. As seen in the attached figure, the tumor-immune interaction is complex and involves multiple immune cells and signaling pathways. We take advantage of whole transcriptome sequencing as a means of interrogating the noted pathways and evaluating the mechanistic results of our viral and vaccine interventions. The resultant large datasets require complex analyses and genomic expertise. We envision that students in the DIMACS program will have an opportunity to work with cell and immunology biologists and those with quantitative genomic expertise.
Project #: DC2016-09
Mentor: Janne Lindqvist, Electrical and Computer Engineering, Rutgers UniversityProject Title: Bitcoin
Bitcoin is a cryptocurrency that is exploding in popularity and has become a darling topic for academic researchers, especially those interested in beefing up its security fundamentals and to improve its technical foundation. But a deeper question lies at the heart of new-age currency that needs to be addressed to validate the amount of energy being expended: what are the usability challenges and barriers to mainstream adoption that stops it from being used en masse? We are looking for students interested in working on potential projects related to human factors in this cryptocurrency including, but not limited to: (i) Bitcoin protocol modifications, (ii) hardware wallets, (iii) usability challenges facing vendors and customers (i.e. plugins and software designed to facilitate transactions), (iv) methodologies for paying with Bitcoin, (v) economic and game theoretic interactions between vendors and miners. Students with backgrounds in CS, Mathematics, EE, and CE are encouraged to apply.
* You must be a U.S. Citizen or Permanent Resident to be eligible for this project.Project #: DC2016-10
Mentor: Janne Lindqvist, Electrical and Computer Engineering, Rutgers UniversityProject Title: Authentication
Mobile authentication, despite myriad research proposals, has stagnated. Many of these proposed alternatives exist only to languish in publication without real-world application or testing -- most never make it out of computing software and on to a device! We are looking for students that are interested in helping us shore up the foundations of mobile authentication by doing rigorous and systematic work to establish clear, useful mobile authentication solutions that address usability, memorability, and security weaknesses inherent to PINs and passwords. Specifically, we want to establish clear metrics for gauging the security space of a solution, the usability benefits, and memorability through user studies. Our current work is focusing mostly on free-form gesture authentication, and we have several related projects going on. Students with backgrounds in CS, Mathematics, CE, and EE are encouraged to apply.
* You must be a U.S. Citizen or Permanent Resident to be eligible for this project.Project #: DC2016-11
Mentor: Janne Lindqvist, Electrical and Computer Engineering, Rutgers UniversityProject Title: Password Security
Passwords are the legacy log-on for desktop environments and have resisted attempts to be replaced. We instead are acknowledging their existence and wish to push forward with better understanding how to help users create and remember better passwords. We are looking for students that are interested in modeling usability and memorability of passwords as created by users and to help solve issues with multiple password interference. The end goal of projects in this space is to increase ways to help users create passwords without unfairly burdening them. Students with backgrounds in CS, CE, EE, and Psychology are encouraged to apply.
* You must be a U.S. Citizen or Permanent Resident to be eligible for this project.
Project #: DC2016-12
Mentor: Janne Lindqvist, Electrical and Computer Engineering, Rutgers UniversityProject Title: Social Network Analysis of Ephemeral Messaging
Ephemeral messaging is on the rise in the United States. The most popular application among young people, for example, is the ephemeral picture messaging application: Snapchat. Yik Yak is now rising as a popular application for millenials in densely populated areas like cities or college campuses. Yik Yak is an anonymous ephemeral location based messaging service that is increasingly being used for short-time bursts of reading, to solicit advice, and even to create connections between people. We are trying to better understand how relationships and social mores develop and propagate in these types of ephemeral messaging services. We are looking for students that are interested in programming modifications of these types of services and to help us conduct user studies and surveys of people who use them. Students with backgrounds in CS, CE, EE, and the social sciences are encouraged to apply. <
Project #: DC2016-13
Mentor: Kim Hirshfield, Rutgers Cancer Institute of New JerseyProject Title: Use of Comprehensive Genomic PRofiling for Personalized Cancer Therapy
Comprehensive genomic profiling of tumors through next-generation sequencing technologies offers the promise of personalized cancer therapy with targeted drugs. However, patient eligibility for genomically-guided drug therapies in the majority of clinical trials is linked to the presence of specific known or characterized genomic alterations. Through an ongoing clinical trial at Rutgers Cancer Institute of New Jersey, we have identified a number of patients whose tumors demonstrate genomic alterations in genes relevant to cancer biology or cancer therapy. While some patients have classic hot-spot mutations or well-characterized mutations, others display genomic findings with predicted altered function or with unknown consequences. We do not yet know how frequently uncharacterized alterations are found in comparison to characterized genomic alterations. Furthermore, for uncharacterized alterations that have been reported in cancer (e.g. in COSMIC or TCGA), we can then use on-line databases to help define which of the uncharacterized genomic alterations are most likely to have functionally-relevant consequences, e.g. through altered downstream gene expression or protein phosphorylation. This approach also offers a new avenue to identify novel mutations for which laboratory-based mutational characterization is most likely to yield data with clinical relevance. By establishing functionality of yet uncharacterized genomic alterations, the potential pool of patients who might benefit from targeted therapy could be increased.
References: Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013; 502:333-9. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, et al. Cancer genome landscapes. Science. 2013; 339(6127): 1546-58. Carter C, Chen S, Isik L, Tyekucheva S, Velculescu VE, Kinzler KW, et al. Cancer-specific High-throughput Annotation of Somatic Mutations: computational prediction of driver missense mutations. Cancer Res 2009; 69: 6660-6667.
Project #: DC2016-14
Mentor: Kellen Myers, Department of Mathematics, SUNY FarmingdaleProject Title: Computational Ramsey Theory
Ramsey theory studies the existence of unavoidable patterns. A mathematical object is colored -- its components are each assigned a particular "color" from a finite set of colors. The question is whether a particular type of structure exists within one of the colored subsets.
For example, if the set {1,2,3,4,5} is colored with colors red and blue, then there is an all-red or all-blue solution to x+y=z. However, if one is limited to {1,2,3,4} the coloring {1,4}=Red and {2,3}=Blue avoids such solutions. We associate to the equation x+y=z the number 5 for this reason: We say the 2-color Rado number for x+y=z is 5.
This project will explore the Rado numbers for other equations, and in some cases with more than two colors. The methods may entail theoretical methods related to the number theory of Diophantine equations, but will also include work on the computational and algorithmic methods used to compute Rado numbers efficiently.
Project #: DC2016-15
Mentor: Kellen Myers, Department of Mathematics, SUNY FarmingdaleProject Title: Epidemiological Modeling
This project will study one or more mathematical models of the dynamics of the spread of disease in various populations. Potential topics include modeling the simultaneous spread of HIV and tuberculosis, analysis of rotavirus immunity in children, and game-theoretic modeling of the spread of disease in populations whose migratory patterns adjust to the presence of disease.
Project #: DC2016-16
Mentor: Christie Nelson, PhD, CCICADA, Rutgers UniversityProject Title: Experimental Designs for Walk Through Metal Detectors at Large Stadium Venues
When utilizing metal detectors at a large venue such as a sports stadium, there are the competing objectives of accuracy of the patron screening and the speed of throughput. This research, carried out in collaboration with large sports venues, analyzes the patron screening method of walk-through metal detectors ("walk-throughs").
Experimental design is the focus of the project, helping large venues better understand the performance of walk-throughs in real outdoor settings. Because of the number of experimental factors to be considered (type of item, location and height of object, orientation, speed of object passing through the machine, walk-through security setting, etc.), designing experiments require a sophisticated design tool called combinatorial experimental design. Experimental designs can focus on various questions, such as: does a walk-through catch each of the pre-specified prohibited test items; can multiple metal test objects can be hidden on a person and tested all at once in different height zones; how does human gait affect detection?
Statistics courses are preferred for this project.
* You must be a U.S. Citizen or Permanent Resident to be eligible for this project.Project #: DC2016-17
Mentor: Wilma Olson, Chemistry & Chemical Biology, Rutgers UniversityProject Title: Dynamics of genome folding: from DNA base pairs to nucleosome arrays and chromosomes
Making the connection between the linear sequence of DNA base pairs and the large-scale organization of the genome requires knowledge of the structure and dynamics of chromatin, the dynamic assembly of DNA and proteins that comprise each chromosome. Our group has developed a multi-scale computational treatment of chromatin that ties the observed folding and dynamics of long genomic fragments to the structures and 3D arrangements of nucleosomes, the spool-like complexes of four duplicated histone proteins and ~150 base pairs of DNA spaced non-randomly along the genome, and the intervening free DNA linkers. Our base-pair-level model of DNA structure and deformability accounts for the global properties of short, well-defined nucleosome arrays, including the preferred sites of nucleosome binding and the communication between DNA elements. We simulate the configurations of long fibers with millions of base pairs in terms of the spatial configurations of nucleosomes collected in the base-pair-level computations. The more coarse-grained, nucleosome-level treatment allows us to study the effects of nucleosome positioning and base sequence on the looping propensities of chromatin fibers comparable in length to the spacing between the physicochemical probes used to characterize the folding of genomic elements in vivo. The build-up of structural components in our treatment of chromatin differs from the many studies of genomic architecture that focus on overall genome organization and thus provides understanding of chromatin folding at the intermediate levels associated with gene regulation and 3D organization and the stepwise processes that underlie genome organization and dynamics. Students will be involved in the generation and/or analysis of simulation data obtained for both small nucleosome arrays modeled at the base-pair level and large-scale chromosome fragments modeled at the nucleosome level.
Project #: DC2016-18
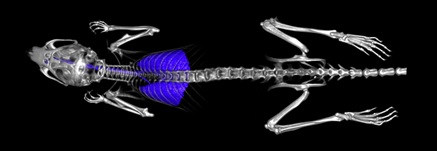
Mentor: Neil Campbell, Rutgers Cancer Institute of New JerseyProject Title: Micro-CT Optimization
The Preclinical Imaging Shared Resource at Rutgers Cancer Institute of New Jersey supports researchers as they study a wide variety of cancers and their potential treatment. One of the modalities that we use is micro-CT (computed tomography), which uses X-rays and computational processing to produce 3D images that can be manipulated in a range of ways. While a single micro-CT scan exposes an animal to an acceptable dose of ionizing radiation, longitudinal studies may, over repeated scans, raise that cumulative dose to clinically relevant levels. Using software, such as Matlab, and micro-CT raw data sets, we hope to identify combinations of acquisition settings and reconstruction algorithms that can allow us to reduce single scan time and exposure to ionizing radiation while providing optimal image quality.
Project #: DC2016-19
Mentor: David Foran, Rutgers Cancer Institute of New JerseyProject Title: High-throughput Analysis of Large Patient Cohorts
A major concentration for the Foran laboratory has been the development of a family of high-throughput, imaging, data-mining and computational tools for classifying malignancies and stratifying biomarker expression signatures from disease onset throughout progression. This work has led to several large-scale, inter-institutional investigative projects in bioinformatics, medical imaging, high-performance computing, computer-assisted diagnostics and therapy planning. Our team is utilizes these resources to conduct high-throughput analysis of large patient cohorts, store and mine large data sets; and determine the relationship between image analysis derived tumor information, gene expression category, patient stratification and clinical outcomes.
Project #: DC2016-20
Mentor: Anand Sarwate, Electrical and Computer Engineering, Rutgers UniversityProject Title: Active learning with label-based costs and communication with feedback
Active learning is a framework for learning decision rules from examples when acquiring examples is expensive. For example, images may need to be annotated by an expert to determine if they are “good” or “bad.” Algorithms for active learning operate sequentially to pick examples to be labeled one at a time this adaptivity can sometimes lead to greater efficiency (though not always). In this project we will look at a variant of this problem where examples cost different amounts to explore, but this cost is revealed only after the decision to explore has been made. This is a new kind of uncertainty because the algorithm has to both learn effectively and keep the overall cost low, and unlike previous models, the cost is not known a priori. This additional uncertainty leads to different tradeoffs. In this project, the REU student will learn about active learning algorithms and explore different methods and heuristics for this new model. Some potentially interesting directions include models for exploring private data, methods to work on higher-dimensional examples, Bayesian frameworks, and connections between this problem and communication with feedback.
Project #: DC2016-21
Mentor: Trefor Williams, Civil and Environmental Engineering, Rutgers UniversityProject Title: Analysis of Railroad Accidents Using Data Analytics, Text Mining and Data Visualization Models
This research will explore various aspects of railroad accidents including truck accidents at highway grade crossings and accidents with hazardous materials. There has been a tremendous growth in oil shipments by rail. We will apply text and data mining to equipment accident reports determine if there are any discernible patterns in the hazardous material accidents that do occur. Initial study of Federal Railroad Administration Databases indicates that large trucks are involved in a disproportionate amount of railroad grade crossing accidents. We will use data visualization to study the integration between various grade crossing factors, such as the type of protection provided and railroad characteristics to better understand grade crossing types that are more likely to have accidents involving large trucks. A knowledge of data mining software like Rapid Miner or Weka would be helpful.
Project #: DC2016-22
Mentor: Midge Cozzens, DIMACS, Rutgers UniversityProject Title: Biodiversity
How do different types of agriculture affect biodiversity and how does biodiversity provide ecosystem services (products of network interactions between species) such as pollination and pest control? Is the role of parasites different from free-living species, especially when considering biodiversity?
References:
i. Algebraic and Discrete Mathematical Methods for Modern Biology, Chapter 2 Food Webs and Graphs, Rena Robeva ed., Elsevier/Academic Press 2015
ii. Owen J. The ecology of garden: the first fifteen years. Cambridge University Press: Cambridge UK. 1991.
iii. Price JM. Stratford-upon-avon: a flora and fauna. Gem Publishing; 2002: Wallingford, UK.
Project #: DC2016-23
Mentor: Midge Cozzens, DIMACS, Rutgers UniversityProject Title: The impact of species extinction on ecosystem services
How do we map ecosystem services onto trophic levels so that food web theory can explain how species extinction or decline leads to reductions in the delivery of these ecosystem services? Can and what would happen if ecosystem service declines before species loss? Why? Are there appropriate models?
References:
i. Algebraic and Discrete Mathematical Methods for Modern Biology, Chapter 2 Food Webs and Graphs, Rena Robeva ed., Elsevier/Academic Press 2015
ii. Kremen C. Managing ecosystem services: what do we need to know about their ecology? Ecology Letters 2005; 8: 468-479.
iii. Dobson A, Allesina S, Lafferty K and Pascual M. The assembly, collapse and restoration of food webs. Transactions of the Royal Society B: Biological Sciences 2009; vol. 364 no. 1574: 16.
Project #: DC2016-24
Mentor: Midge Cozzens, DIMACS, Rutgers UniversityProject Title: Invasive species in food webs
Invasive species pose significant threats to habitats and biodiversity. Studies to date consider the effects of alien species at a single trophic level, yet linked extinctions and indirect effects are not recognized from this approach. Food webs can often identify these indirect effects and the impact of the addition of an alien species into the network, such as competition due to shared enemies. The removal of an invasive species is a priority for conservationists and the public, so these species provide opportunities for manipulative field experiments testing the removal of a node from a food web. Consider the effects of an invasive species on the whole food web.
References:
i. Algebraic and Discrete Mathematical Methods for Modern Biology, Chapter 2 Food Webs and Graphs, Rena Robeva ed., Elsevier/Academic Press 2015
ii. Memmott J. Food webs: a ladder for picking strawberries or a practical tool for practical problems? Transactions of the Royal Society B: Biological Sciences 2009; 364 no. 1574: 1693-1699.
Project #: DC2016-25
Mentor: Midge Cozzens, DIMACS, Rutgers UniversityProject Title: Competition graphs and food webs
What are the ecological characteristics of food webs that seem to lead to their competition graphs being interval graphs? “Most directed graphs” do not have interval graph competition graphs, yet statistically most actual food webs have interval competition graphs. (BIG unsolved problem mentioned by Cohen but no answers to date: Cohen JE, Komlos J,and Mueller M. The Probability of an Interval Graph and Why it Matters. Proc. Symposium on Pure Math 1979; 34: 97-115.) In this project, we will analyze the properties of competition graphs to better understand the underlying food web (directed graph). Consider the following questions: Can we characterize the directed graphs whose corresponding competition graphs are interval graphs? This is a fundamental open question in applied graph theory. Indeed there is no forbidden list of digraphs (finite or infinite) such that when these digraphs are excluded, one automatically has a competition graph which is an interval graph.
References:
i. Algebraic and Discrete Mathematical Methods for Modern Biology, Chapter 2 Food Webs and Graphs, Rena Robeva ed., Elsevier/Academic Press 2015
ii. Opsut R. On the competition number of a graph. SIAM Journal of Algebraic and Discrete Methods 1982; 3: 470-478.
iii. Cho HH and Kim SR. A class of acyclic digraphs with interval competition graphs. Discrete Applied Mathematics 2005; 148: 171-180.
iv. MacKay B, Schweitzer P and Schweitzer P. Competition numbers, quasi-line graphs, and holes. SIAM J. of Discrete Math 2014; 28(1): 77-91.
Project #: DC2016-26
Mentor: Midge Cozzens, DIMACS, Rutgers UniversityProject Title: Dominating sets in graphs
A set of vertices is a dominating set in a graph if every vertex not in the dominating set is adjacent to one or more vertices in the dominating set. A dominating clique is a dominating set that is a clique. Cozzens and Kelleher some years ago gave a forbidden subgraph characterization for graphs that contain a dominating clique. It had long been known when a graph had a single dominating vertex or a dominating edge. In addition, they gave a forbidden subgraph characterization for graphs that contain a dominating K3 or a P3. Graphs with dominating cliques are closely related to split graphs.
Questions:
1. Is there a forbidden subgraph characterization of graphs which have a connected dominating set of size 4? Note that the only connected graphs on 3 vertices are K3 and P3.
2. Are there polynomial time algorithms for locating a minimum dominating clique in a clique dominated graph? What other parameters are easy to calculate for clique dominated graphs?
3. Is there a forbidden subgraph characterization of graphs that have a dominating induced path?
References:
i. Kulli, V.R.. Theory of Domination in Graphs, Vishwa International Publications, India 2010.
ii. Cozzens, M.B. and Kelleher, L. L.. Dominating Cliques in Graphs, Discrete Math 86 (1990). 303-308.
Project #: DC2016-27
Mentor: Erica Ashe and Robert Kopp, Department of Earth and Planetary Sciences, Rutgers UniversityProject Title: Bayesian Statistical Modeling: Gaussian Process Regression Applied to Sea-Level Data
A Gaussian process defines a distribution over functions, which can be used for Bayesian regression. GPs are fully defined by their mean function and covariance function, or kernel. We use GP regression to model relative sea level as a spatiotemporal Gaussian field. This project will involve adapting current models to new sea-level data and exploring the possibility of non-Gaussian likelihoods. Some background in programming is encouraged.
References:
Gaussian Processes for Machine Learning, Rasmussen and Williams, MIT
Press, http://www.gaussianprocess.org/
Holocene Relative Sea-Level Changes from Near-, Intermediate-, and Far- Field Locations, Khan et. al., http://link.springer.com/article/10.1007%2Fs40641-015-0029-z
Project #: DC2016-28
Mentor: John Kolassa, Department of Statistics, Rutgers UniversityProject Title: Applications of Bootstrap Methods
Statisticians have long proposed methods for estimating quantities of conceptual interest arising in empirical studies. These quantities include mean effects of treatments, correlations, and measures of association between variables of interest. When statisticians estimate such quantities, they generally want to describe the range of plausible values for such a quantity. This range is generally called a confidence interval. Constructing such intervals generally relies on assumptions about the probability structure generating the underlying data set. A set of modern computationally-intensive techniques, collectively known as the bootstrap, uses only data contained in the data set to construct such intervals. I propose numerically surveying the application of the various bootstrap methods to various statistical estimators, arising from various underlying distributions, to assess and compare the accuracy of these methods.
Project #: DC2016-29
Mentor: James Abello, DIMACS, Rutgers UniversityProject Title: Visualization of Time-Varying Graphs
ASK-GraphView is a node-link-based graph visualization system that allows clustering and interactive navigation of large graphs [1]. The system constructs a hierarchy on an arbitrary, weighted, undirected input graph using a series of increasingly sophisticated clustering algorithms. Building on this earlier work, this project will address the problem of visualizing graphs that are being collected in a streaming fashion. A key challenge is to identify graph sub-structures and statistics on them that can be used as the basis to depict evolution of the graph through time [2]. Another goal is to devise a visualization algorithm that produces a layout that is stable over time, thus preserving a user's mental map.
References:
[1] Abello J, Ham FV, Krishnan N, "AskGraphView: A Large Graph Visualization System", IEEE Transactions in Visualization and Computer Graphics, 12(5) (2006), 669-676.
[2] Hu Y, Kobourov S, and Veeramoni S, "Embedding, Clustering and Coloring for Dynamic Maps", Proceedings of IEEE Pacific Visualization Symposium, (2012).
Project #: DC2016-30
Mentor: James Abello, DIMACS, Rutgers UniversityProject Title: Mapping the OEIS (https://oeis.org)*
The Online Encyclopedia of Integer Sequences (OEIS) is a marvelous resource initiated in 1967 by mathematician Neil Sloane (neilsloane.com). It currently has over 260,000 integer sequences, attracting about 9 million hits a month. Most people use the OEIS to get information about a particular integer sequence. If a sequence is not in the database, the user may submit it. It is then analyzed by OEIS editors and if found “interesting”, it is added to the repository together with comments about the provenance of the sequence and references to where the sequence or related sequences appear. This dataset is publicly available. It has generated tantalizing open ended research questions, spawned many mathematical discoveries, and has been cited thousands of times. The main data entities here are integer sequences. At the time of this writing, OEIS does not have an overall classification scheme for the sequences, or an operational definition of sequence “similarity”. This suggests that the only feasible mechanism for researchers to learn about the dataset structure is by enhancing the current user exploration mechanisms, and classifying interactively the users exploration “trails” and findings. The picture below represents one OEIS topology defined by DIMACS 2015 REU students [5].

One of our challenges is to use adaptive local exploration to navigate and create or refine topologies for the OEIS in consultation with faculty and students in mathematics, physics, engineering,
and OEIS’s founder Neil Sloane and OEIS’s editors. The essential elements of the exploration are:
i. an underlying topology defined by a mathematical formalism which is not explicitly stored
ii. global statistics of the co-occurrence of appearing patterns derived from conjecture descriptions
iii. a data contraction mechanism
iv. the ability to navigate locally and on demand according to some underlying topology
v. a mechanism to “teleport” from an equivalence class (i.e. a subgraph pattern ) to a “similar” pattern that is topologically distant.
References:
[1] J. Abello, F. Van Ham, N. Krishnan, ASK-Graph View: A Large Scale Graph Visualization System. In IEEE Transactions on Visualization and Computer Graphics, Vol. 12, No. 5, Sept/Oct. 2006.
[2] N.Bikakis.Towards Scalable Visual Exploration of Very Large RDFGraphs. In arXiv:1506.04333, 2015.
[3] L. Nachmanson, GraphMaps: Browsing Large Graphs as Interactive Maps. In arXiv:1506.06745, 2015.
[4] L. Shi. Hierarchical Focus+Context HeterogeneousNetwork
Visualization. In IEEE Pacific Visualization Symposium
(PacificVis’14), 2014.
[5] J. Abello, B. Mariscal, D. Mahwritter, K. Sun, OEIS Exploration, DIMACS REU 2015.
Project #: DC2016-31
Mentor: James Abello, DIMACS, Rutgers UniversityProject Title: Mapping PubChem
Existing chemical data bases contain information about millions of known molecules. The information includes molecule classification, chemical structure, and patent information. PubChem is one such data repository with descriptions of over 8 million molecules. In this project, one of the aims is to create a PubChem hierarchical graph topology that can be queried, visualized, navigated, and annotated with user and curators findings. The processing will use a combination of MapReduce and DbMongo using the *Graph Cards abstraction developed in previous DIMACS REU years.
Web Resources:
Public molecule database: http://pubchem.ncbi.nlm.nih.gov/#
Information about MapReduce: http://static.googleusercontent.com/media/research.google.com/es//archive/mapreduce-osdi04.pdf
Graph visualization: http://www.mgvis.com/Papers/ASK-Graphview3.pdf
*Graph Cards: J. Abello, David Desimone, M. Sumida, DIMACS REU 2014, 2015
Project #: DC2016-32
Mentor: Muthu Muthukrishnan, Computer Science, Rutgers UniversityProject Title: How to Analyze Data Privately despite Breakins?
Given the log of people entering and leaving a building, suppose one wants to estimate how many different people are in the building at any time. Can this be done privately? Differential Privacy provides such a method. But suppose attackers sneak into the system that does this calculation. Can the analysis be still kept private? We have developed algorithms for this problem and many others of similar ilk. This project during the summer is to explore these algorithms and possibly extend them. Such algorithms are of great importance in a world where people carry many personal devices and keep valuable information including finances, friends and health in these devices.
Project #: DC2016-33
Mentor: Muthu Muthukrishnan, Computer Science, Rutgers UniversityProject Title: How to Quantify Inflation in Citations (and Grades)?
Different research areas have different approaches to citing others' work. This leads to citation metrics that are difficult to interpret across research areas. The goal of this project is to quantify and correct for this effect. In particular, we will pursue a random walk or page rank style approach to this problem, and either implement and test existing superfast algorithms or extend them suitably.
Project #: DC2016-34
Mentor: Christopher Woodward (Math REU Coordinator), Mathematics, Rutgers UniversityProject Title: Projects in Pure Math
Several projects will be run in 2016 by members of the pure math faculty. Past topics have included Schubert calculus (Buch) Ricci flow(Song) mathematical physics and general relativity (Soffer) and mathematical Yang-Mills theory (Woodward).
Applicants interested in a project in pure mathematics should list this project in their application; further details will be worked out in the Spring.
Project #: DC2016-35
Mentor: James Abello, DIMACS, and Yana Bromberg, Department of Biochemistry and Microbiology, Rutgers UniversityProject Title: Proteins Fold Similarity
Energy is an organism's most basic need. Redox reactions, i.e. electron transfer reactions, are necessary to fulfill the energy requirements of all life forms. The ability to carry out redox reactions must have been among the first functionalities acquired by early life forms. Understanding the evolution of biological redox machinery, i.e. the molecular pathways of interacting proteins/enzymes, can shine light on the history of life, as we know it. Redox enzymes often use metals and metal-containing ligands. The evolution of redox abilities can thus be mapped through the analysis of structural similarity of metal-binding protein folds, i.e. the active components of redox enzymes.
REU Students involved in this project will utilize computational and mathematical techniques to perform network analysis on metal-binding protein folds similarity graphs to determine relationships amongst proteins. Students will implement graph decomposition and network flow based algorithms to visualize protein folds similarity in creative and captivating ways. The overall goal is to enhance our current understanding of whether metal binding folds in non-redox proteins may have come from redox-fold ancestors. This information will then be used to build a variety of corresponding protein evolutionary trees.
References:
J. Abello and F. Queyroi, Fixed Points of Degree
Peeling, ASONAM
'13 Proceedings of the 2013 IEEE/ACM International Conference on
Advances in Social Networks Analysis and Mining, pages 256-263, doi
10.1145/2492517.2492543.
Harel, A., Bromberg, Y., Falkowski, P.G., Bhattacharya, D. (2014) Evolutionary history of redox metal-binding domains across the tree of life. Proc Natl Acad Sci U S A 111(19):7042-7047..
Project #: DC2016-36
Mentor: Jingjin Yu, Department of Computer Science, Rutgers UniversityProject Title: Sharp Transitions of Collision Behavior in Multi-Robot Systems
Algorithms for multi-robot path planning are fundamental building blocks for enabling fully autonomous robots, with applications covering collaborative manufacturing, autonomous driving (e.g., autonomous cars), warehousing (e.g., Kiva systems), autonomous aerial delivery (e.g., Amazon air and Google X-wing), and so on. The problem of planning non-colliding (and ideally optimal) paths for a large number of robots sharing the same workspace is highly challenging. Indeed, such problems have been shown to be NP-hard in discrete settings and PSPACE-hard in continuous settings. On the other hand, when the density of robots is low, planning paths is usually easy using variants of A* search.
The two sides of the multi-robot path planning problem seem to point to a "sharp transition" in algorithm efficiency as robot density in the workspace increases. Such transitions are actually rather common, for example, the connectivity of random geometric graphs. This project seeks to understand such behavior through a combination of simulation study, algorithm development, and theoretical analysis. We wish to either confirm or reject the the sharp transition hypothesis, and exploit the gained insight to develop more efficient multi-robot path planning algorithms.
References:
J. Yu. Intractability of Optimal Multi-Robot Path Planning on Planar Graphs. IEEE Robotics and Automation Letters
J. Yu and Daniela Rus. Pebble Motion on Graphs with Rotations: Efficient Feasibility Tests and Planning Algorithms. In Algorithmic Foundations of Robotics XI, Springer Tracts in Advanced Robotics (STAR), Vol 107, page(s): 729-746, 2015. Video: https://youtu.be/eg3tKIv2eiU
M. Penrose, Random Geometric Graphs, ser. Oxford Studies in Probability. London: Oxford University Press, 2003.