March 2018
The energy (and entropy) level at DIMACS increases along with the summer temperatures as we welcome students participating in our Research Experiences for Undergraduates (REU) program. The DIMACS REU program combines an NSF REU site with several other programs to bring a large and energetic cohort of undergraduates to Rutgers each year. In 2017, DIMACS hosted 30 students: 11 were supported by the site grant, two by NSF REU supplements, four by the Rutgers Mathematics Department, three by CCICADA (the homeland security center based at DIMACS), two by the US Air Force Academy in association with CCICADA, one by the Rutgers Project SUPER program, six by the DIMATIA Center at Charles University in Prague, and one by the Thai-American Association ATPAC.
We present a few highlights from the 2017 REU here.
Andrea Burns (Tulane University) worked with ECE professor Waheed Bajwa on a project that showed the advantage of using multimodal data in the classification of liquids using machine learning and image processing.  Multimodal data have made a huge impact on the field of machine learning because they better represent the way humans learn to perform tasks. One example of multimodal data is that of hyperspectral imaging (HSI), where multimodality corresponds to different spectral ranges. Burns created and curated a Hyperspectral Liquid 12-Band dataset (HyL12) by recording images of various liquids through different cameras, capturing images at different wavelengths. For example, the pictures at right show a bottle seen through visible (left) and UV (right) light cameras. Burns's work uses information from many spectral bands to classify the liquid contained in the bottle. Burns used Support Vector Machine methods to show the increased accuracy of her method in correctly classifying unknown liquids. This work has the potential to drastically reduce screening time in airport security or at any large event where liquids may pose security concerns. Burns and Bajwa’s research has been accepted for publication in the SPIE Defense + Commercial Sensing Conference, where Burns will present next month in Orlando.
Multimodal data have made a huge impact on the field of machine learning because they better represent the way humans learn to perform tasks. One example of multimodal data is that of hyperspectral imaging (HSI), where multimodality corresponds to different spectral ranges. Burns created and curated a Hyperspectral Liquid 12-Band dataset (HyL12) by recording images of various liquids through different cameras, capturing images at different wavelengths. For example, the pictures at right show a bottle seen through visible (left) and UV (right) light cameras. Burns's work uses information from many spectral bands to classify the liquid contained in the bottle. Burns used Support Vector Machine methods to show the increased accuracy of her method in correctly classifying unknown liquids. This work has the potential to drastically reduce screening time in airport security or at any large event where liquids may pose security concerns. Burns and Bajwa’s research has been accepted for publication in the SPIE Defense + Commercial Sensing Conference, where Burns will present next month in Orlando.
Ondrej Maxian (Case Western Reserve University) worked with Chemistry professor Wilma Olson and her graduate student Stefjord Toddoli to develop mathematical tools for the analysis of chromatin. The  packing of DNA into nucleosome core particles (NCPs) is vital to the field of epigenetics and gene expression. Until now, the packing of NCPs has been studied physically via X-rays and cryogenic electron microscopy (cryo-EM), with few mathematical tools available. Maxian developed tools based on computational geometry to quantitatively evaluate the strength of interaction between two nucleosomes on their faces and sides, as well as to give biological meaning to the regions of interaction. The method was applied to X-ray crystal and cryo-EM structures, largely confirming existing observations while also developing new insights into some atypical structures. Maxian also used this algorithm to analyze configuration outputs from Markov Chain Monte Carlo (MCMC) simulations, which were found to closely resemble those from cryo-EM and X-ray. Maxian presented his work at the American Chemical Society (ACS) National Meeting earlier this month.
packing of DNA into nucleosome core particles (NCPs) is vital to the field of epigenetics and gene expression. Until now, the packing of NCPs has been studied physically via X-rays and cryogenic electron microscopy (cryo-EM), with few mathematical tools available. Maxian developed tools based on computational geometry to quantitatively evaluate the strength of interaction between two nucleosomes on their faces and sides, as well as to give biological meaning to the regions of interaction. The method was applied to X-ray crystal and cryo-EM structures, largely confirming existing observations while also developing new insights into some atypical structures. Maxian also used this algorithm to analyze configuration outputs from Markov Chain Monte Carlo (MCMC) simulations, which were found to closely resemble those from cryo-EM and X-ray. Maxian presented his work at the American Chemical Society (ACS) National Meeting earlier this month.
Simon Bird (Rutgers University) was a co-author in a recent publication in the journal JCO Precision Oncology. Part of this paper was based on Bird’s REU research with Hossein Khiabanian, a professor at the Cancer Institute of New Jersey. Bird developed a computational model using simulations and statistical analysis to determine the probability—given biopsy results for a tumor sample—that a mutation is either hereditary or that it developed during tumor progression. The extensive tumor simulations were able to show the kinds of tumors for which mutational status can be inferred. The key factor identified was the ploidy of the sample, defined as the number of chromosomal copies at the mutation’s locus. Given a known ploidy, the model has an impressive 95% accuracy in 68% of cases. Otherwise, the correct model can be predicted only 11% of the time, while no conclusion can be reached for the other cases. This implies that in many scenarios, doctors do not require additional tumor sequencing in order to know the mutational status. This can be useful for both treatment and preventive purposes.
Oncology. Part of this paper was based on Bird’s REU research with Hossein Khiabanian, a professor at the Cancer Institute of New Jersey. Bird developed a computational model using simulations and statistical analysis to determine the probability—given biopsy results for a tumor sample—that a mutation is either hereditary or that it developed during tumor progression. The extensive tumor simulations were able to show the kinds of tumors for which mutational status can be inferred. The key factor identified was the ploidy of the sample, defined as the number of chromosomal copies at the mutation’s locus. Given a known ploidy, the model has an impressive 95% accuracy in 68% of cases. Otherwise, the correct model can be predicted only 11% of the time, while no conclusion can be reached for the other cases. This implies that in many scenarios, doctors do not require additional tumor sequencing in order to know the mutational status. This can be useful for both treatment and preventive purposes.
Aaron Zhang (Brown University) worked with Computer Science professor Desheng Zhang and his graduate student Zhihan Fang to develop FineTravel, a model for travel-time estimation based on 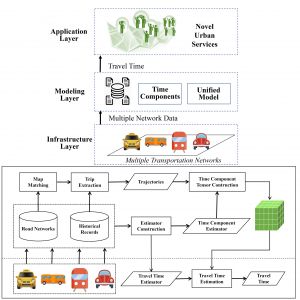 multiple transportation networks. Most existing work on travel-time estimation has focused on estimating riding time for a single mode of transportation. However, passengers often use different modes of transportation, e.g. subways and buses, and a significant portion of travel time is spent walking and waiting. The FineTravel model estimates travel time with fine-grained time components: waiting time, walking time, and riding time. In the model, travel time is divided into components by passenger status and transportation modality. Estimators for each component are constructed from historical data. The performance of the travel-time estimation was evaluated on large-scale real-world data from multiple sources in four transportation networks: subway, taxis, buses, and private vehicles. This model was found to be more accurate than baseline estimates in all modalities, highlighting the potential of this work to significantly improve urban transportation planning.
multiple transportation networks. Most existing work on travel-time estimation has focused on estimating riding time for a single mode of transportation. However, passengers often use different modes of transportation, e.g. subways and buses, and a significant portion of travel time is spent walking and waiting. The FineTravel model estimates travel time with fine-grained time components: waiting time, walking time, and riding time. In the model, travel time is divided into components by passenger status and transportation modality. Estimators for each component are constructed from historical data. The performance of the travel-time estimation was evaluated on large-scale real-world data from multiple sources in four transportation networks: subway, taxis, buses, and private vehicles. This model was found to be more accurate than baseline estimates in all modalities, highlighting the potential of this work to significantly improve urban transportation planning.
All of our 2017 REU students demonstrated impressive skills, determination, and hard work during their summer experience, and we look forward to hearing and sharing more about their future endeavors. Read about all of the 2017 REU students' projects here.
Printable version of this story: [PDF]
References
A. Burns and W.U. Bajwa. “Multispectral imaging for improved liquid classification in security sensor systems”. Accepted for publication in SPIE Defense + Commercial Sensing Conference (2018).
O. Maxian, S. Todolli, and W.K. Olson “Using Mathematical Tools to Characterize Chromatin Geometries in Crystal, Cryo-EM and Simulated Structures”. Oral presentation at the American Chemical Society Spring Meeting, March 18-22, 2018, New Orleans LA.
H. Khiabanian, K.M. Hirshfield, M. Goldfinger, S. Bird, M. Stein, J. Aisner, D. Toppmeyer, S. Wong, N. Chan, K. Dhar, J. Gheeya, H. Vig, M. Hadigol, D. Pavlick, S. Ansari, S. Ali, B. Xia, L. Rodriguez-Rodriguez, and S. Ganesan. “Inference of germline mutational status and evaluation of loss of heterozygosity in high-depth tumor-only sequencing data.” JCO Precision Oncology, 1-15, doi:10.1200/PO.17.00148 (2018).
Z. Fang, A. Zhang, and D. Zhang. “FineTravel: Fine-Grained Travel Time Estimation for Multiple Transportation Networks.” In submission.



